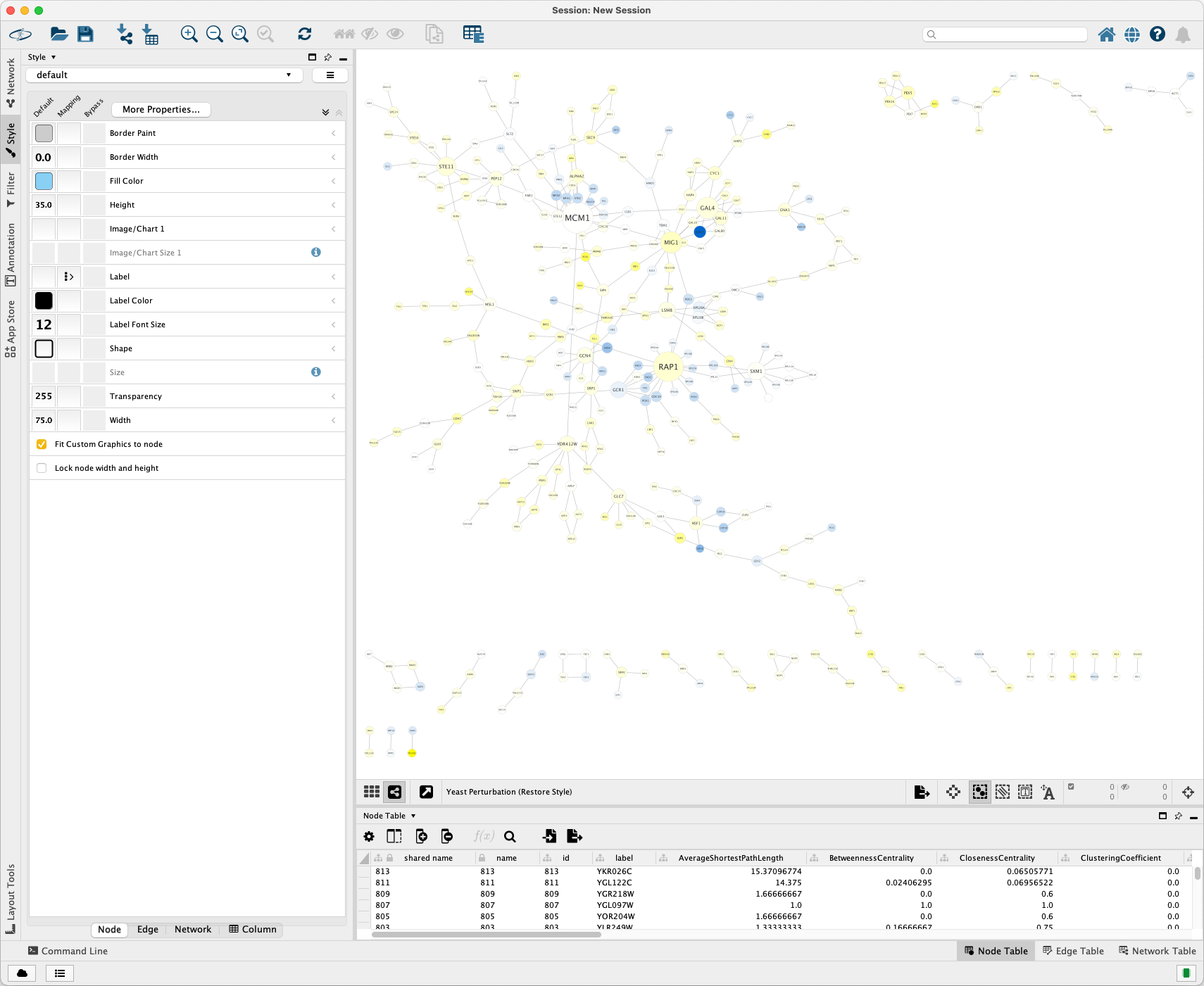
Examples¶
Sample data¶
Open Cytoscape and select the Yeast Perturbation sample data.
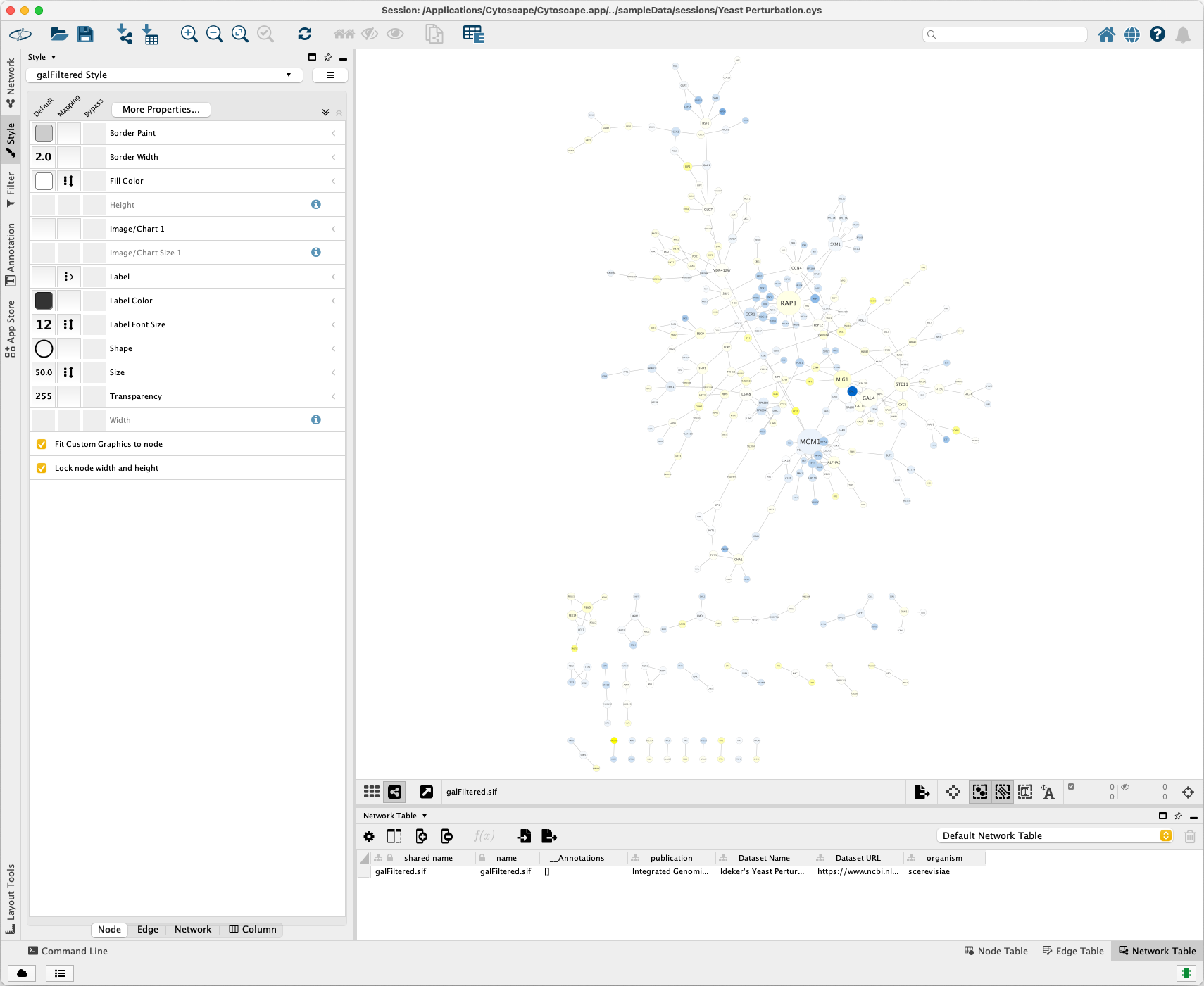
Click File > Export > Network to File... to export it as an XGMML format file.
The exported yeast_perturbation.xgmml file is stored in this directory of the repository.
Reader¶
Run the following code to read the XGMML file with NetworkxXGMMLReader:
from xgmml.reader.networkx_xgmml_reader import NetworkxXGMMLReader
xgmml_reader = NetworkxXGMMLReader()
yeast_perturbation_graph = xgmml_reader.read('yeast_perturbation.xgmml')
Run the following code to view the basic information of the graph:
yeast_perturbation_graph.is_directed()
# True
len(yeast_perturbation_graph.nodes)
# 330
len(yeast_perturbation_graph.edges)
# 359
Writer¶
Run the following code to write the graph into an XGMML file with NetworkxXGMMLWriter:
from xgmml.writer.networkx_xgmml_writer import NetworkxXGMMLWriter
xgmml_writer = NetworkxXGMMLWriter()
xgmml_writer.write(
yeast_perturbation_graph,
'yeast_perturbation.xgmml',
graph_label='Yeast Perturbation (Default)',
)
Import it into Cytoscape. The visualization is shown as below:
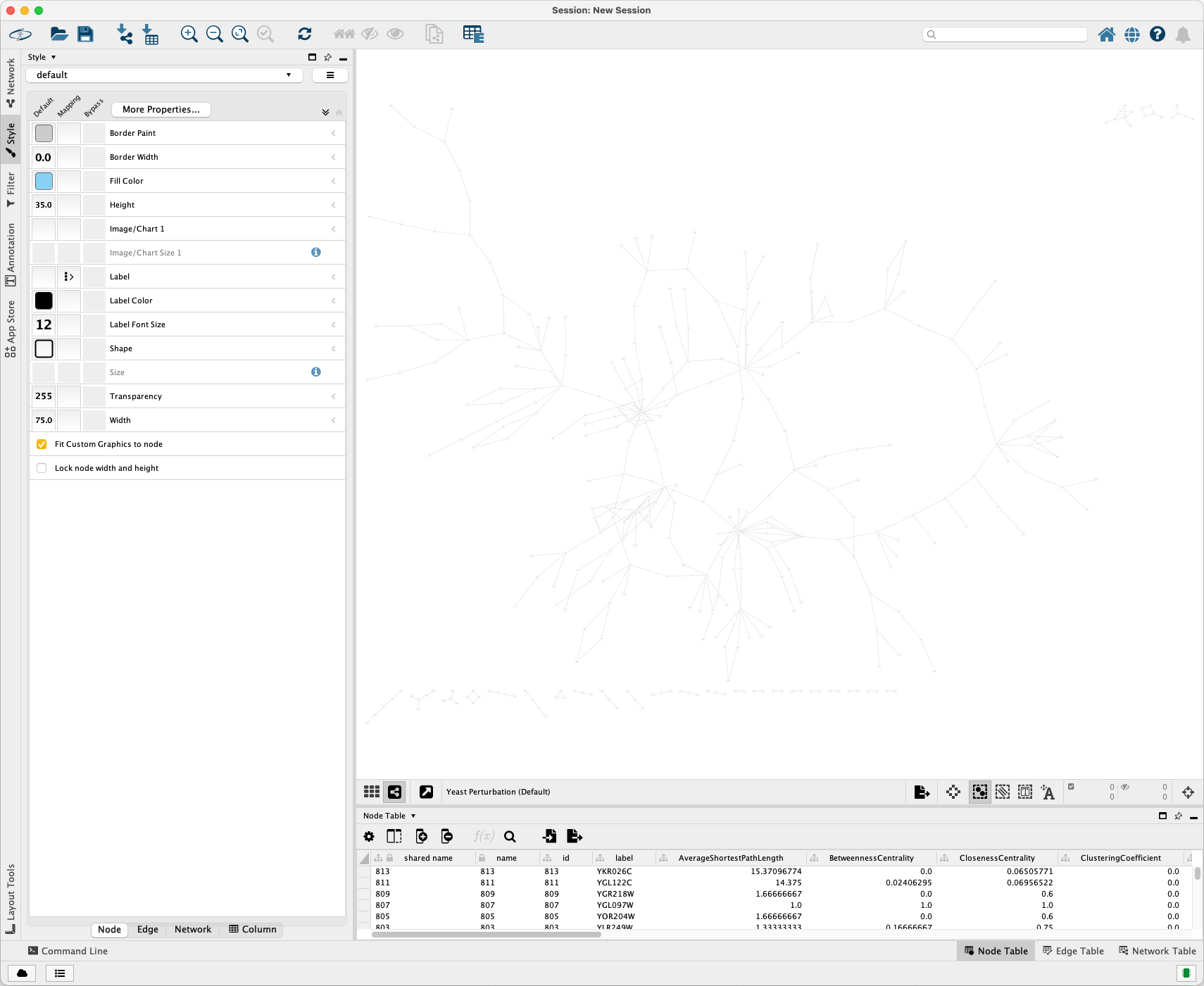
Run the following code to create a similar style to the original XGMML file and write it into an XGMML file:
import matplotlib as mpl
from xgmml.writer.networkx_xgmml_writer import NetworkxXGMMLWriter
from xgmml.style.style_mapper import *
class StyleNodeSizeMapper(StyleNodeSizeDefaultContinuousMapper):
def __init__(self, values, **kwargs):
super(StyleNodeSizeMapper, self).__init__(values, **kwargs)
self._min_size = 40
self._max_size = 150
class StyleNodeLabelFontSizeMapper(StyleFontSizeDefaultContinuousMapper):
def __init__(self, values, **kwargs):
super(StyleNodeLabelFontSizeMapper, self).__init__(values, **kwargs)
self._min_size = 10
self._max_size = 40
class StyleNodeFillColorMapper(StyleColorContinuousMapper):
def __init__(self, values, **kwargs):
colors = ['#0066CC', '#FFFFFF', '#FFFF00']
colormap = mpl.colors.LinearSegmentedColormap.from_list('custom', colors)
super(StyleNodeFillColorMapper, self).__init__(
values, colormap=colormap, **kwargs
)
xgmml_writer = NetworkxXGMMLWriter()
xgmml_writer.write(
yeast_perturbation_graph,
'yeast_perturbation.xgmml',
graph_label='Yeast Perturbation (Restore Style)',
node_label_key='COMMON',
node_width_key='degree.layout',
node_width_mapper=StyleNodeSizeMapper,
node_height_key='degree.layout',
node_height_mapper=StyleNodeSizeMapper,
node_label_font_size_key='Degree',
node_label_font_size_mapper=StyleNodeLabelFontSizeMapper,
node_fill_color_key='gal1RGexp',
node_fill_color_mapper=StyleNodeFillColorMapper,
)
Import it into Cytoscape. The visualization is show as below:
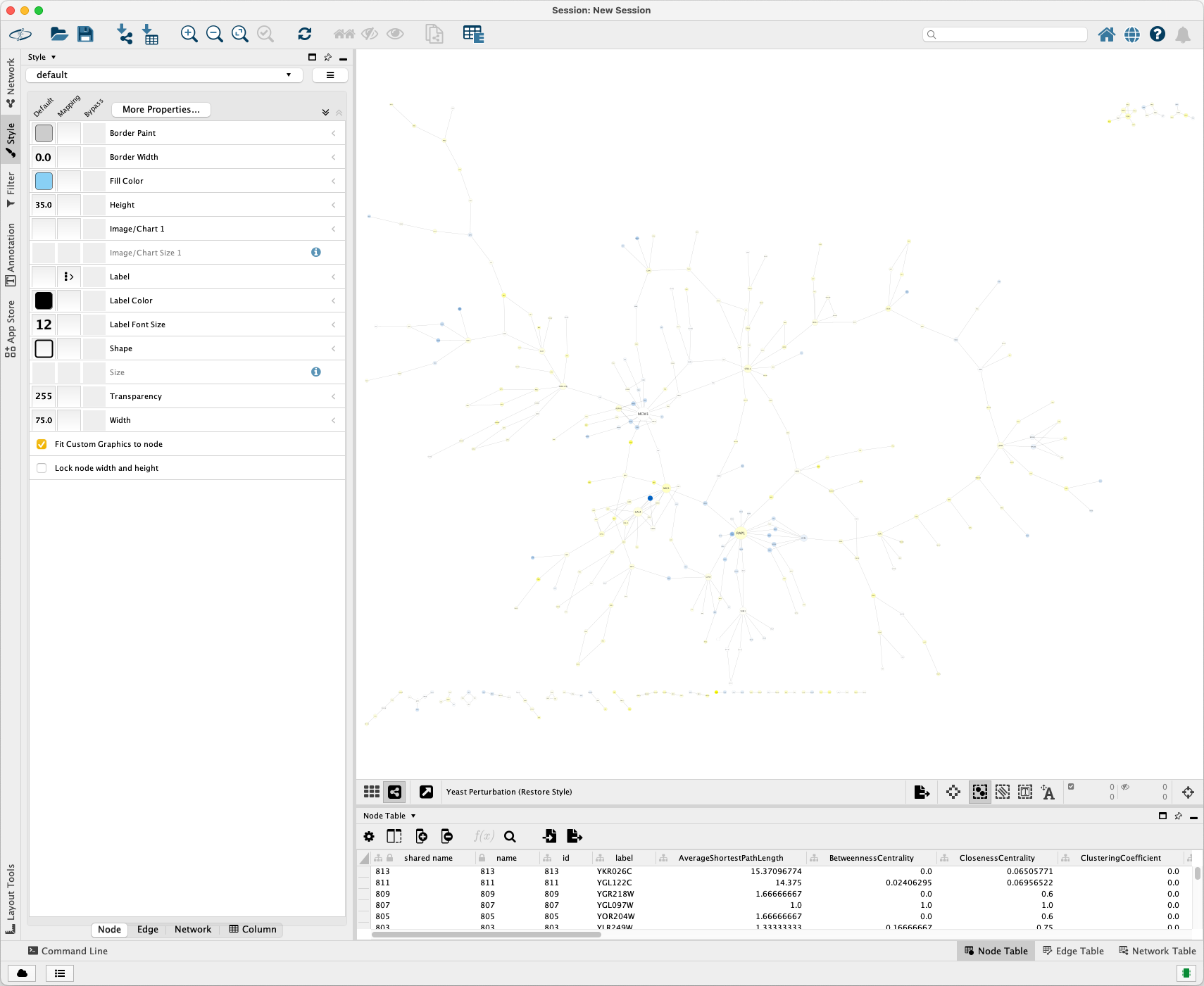
Click Layout > Prefuse Force Directed Layout to get a better layout.